
Novel microscope and software allow imaging at different scales, tracking biological processes over long time periods

A new “self-driving” microscope developed by UT Southwestern Medical Center researchers solves two fundamental challenges that have long plagued microscopy: first, imaging living cells or organisms at dramatically different scales, and second, following a specific structure or area of interest over long periods of time. This innovation, detailed in Nature Methods, is already making observations that have not been possible with conventional methods.

Reto Fiolka, Ph.D., (left) is Associate Professor in the Lyda Hill Department of Bioinformatics and of Cell Biology at UT Southwestern. Stephan Daetwyler, Ph.D., is Instructor in the Hill Department of Bioinformatics.
“Our work demonstrates a significant advancement in integrated bioimaging, enabling long-term observation of biological dynamics across scales from cellular to systemic levels to ultimately better understand developmental and disease processes,” said Reto Fiolka, Ph.D., Associate Professor in the Lyda Hill Department of Bioinformatics and of Cell Biology at UT Southwestern. Dr. Fiolka co-led development of the new microscope with Stephan Daetwyler, Ph.D., Instructor in the Lyda Hill Department of Bioinformatics.
For two decades, Dr. Daetwyler explained, researchers have made a wealth of biological discoveries using a technique called light-sheet microscopy, in which a thin plane of light excites fluorescent probes added to a sample to tag specific structures. Because the light causes these probes to glow, researchers could easily find and image the tagged structures, allowing for a deeper comprehension of their role in health and disease.
However, Dr. Daetwyler said, light-sheet microscopy comes with a significant drawback: The higher the imaging resolution, the smaller the area researchers can image. Thus, scientists have had to choose which resolution and field of view they required before starting an experiment. Therefore, they could either image living whole organisms at low resolution, or a few cells and their internal structures at high resolution. This limitation has significantly hindered understanding of biological processes, since most of them happen on the cellular, tissue, and organismal levels, Dr. Daetwyler said.
To address this challenge, Drs. Fiolka and Daetwyler and their colleagues combined hardware that performs low-resolution light-sheet microscopy, known as multidirectional selective plane illumination microscopy (mSPIM), with hardware that performs a type of high-resolution light-sheet microscopy called axially swept light-sheet microscopy (ASLM, developed at UTSW in 2015). To switch between the two modalities within a second, the team created a novel custom microscope hardware control software.
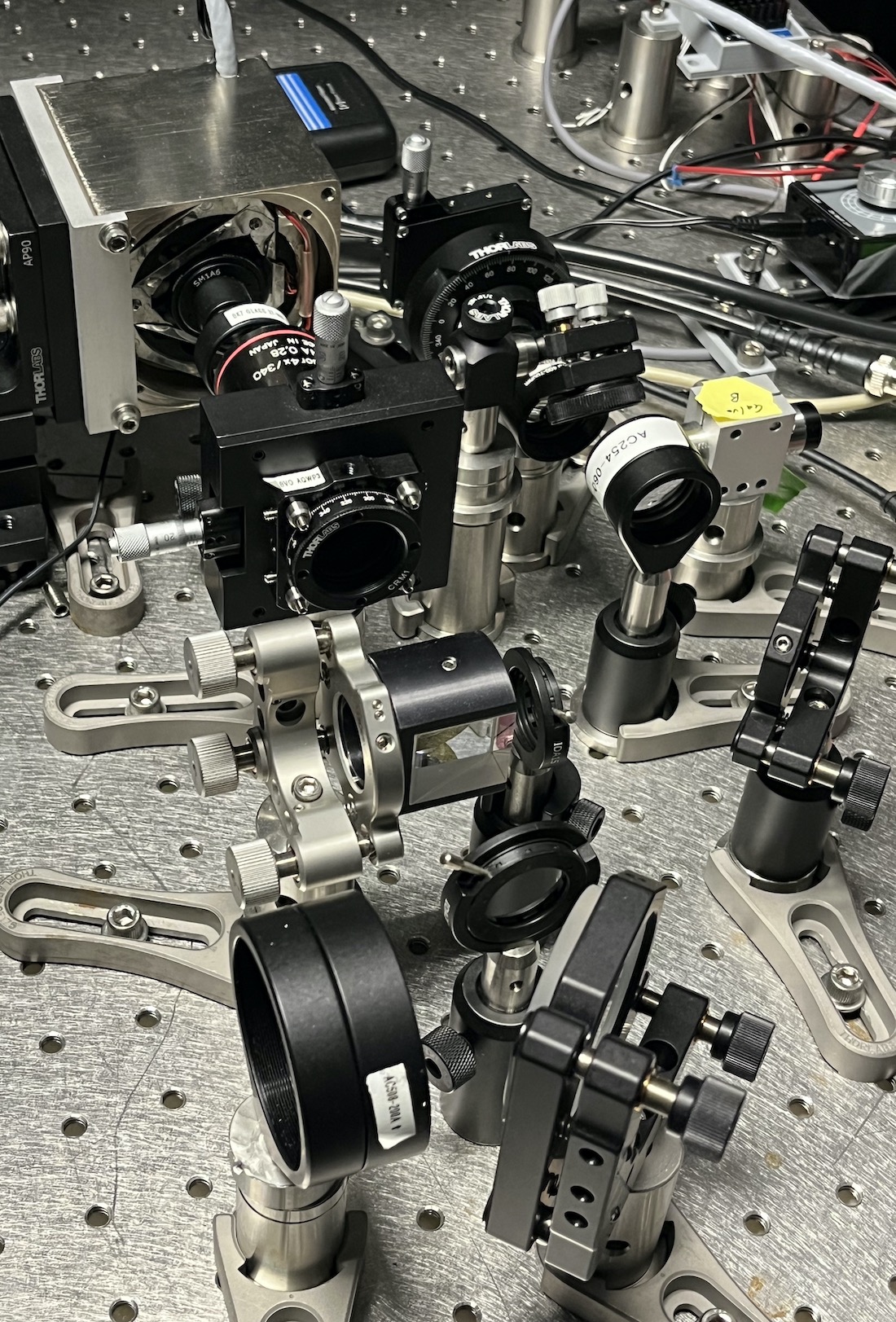
In this part of the microscope, the high-resolution light-sheet is pivoted back and forth for making the axially swept light-sheet modality for high-resolution imaging, and the low- and high-resolution light-sheets are combined onto the same illumination path.
Another challenge with light-sheet microscopy has been tracking structures over time, Dr. Fiolka added. Most biological processes are dynamic, he explained. Organisms grow, and cells move and multiply, requiring human intervention to frequently redirect the microscope’s field of view – a tedious task when imaging biological processes that take place over hours or days.
To find a solution, the researchers incorporated a tracking mode in the new microscope’s control software to imbue it with a “self-driving” function, allowing the microscope’s field of view to follow a specific region of interest – which users register during an initialization step – for hours or even days.
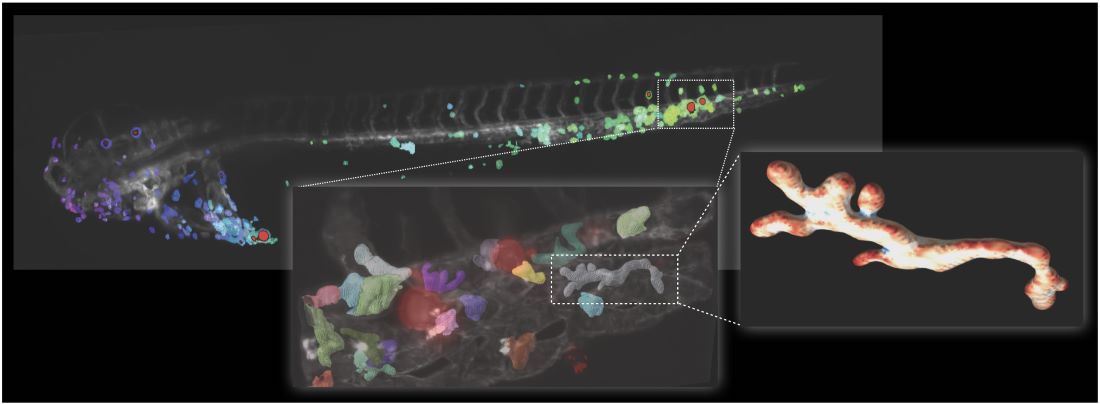
As proof of principle, the researchers imaged human cancer cells injected into zebrafish larvae, transparent organisms a few millimeters long that are frequently used as biological models for cancer research. The researchers saw that osteosarcoma cells – a type of bone cancer – were readily attacked and cleared by immune cells called macrophages. However, skin cancer cells were not cleared from the organism, despite close interactions with the immune cells. Zooming in on the macrophages, the team observed changes in shape that correlated with their current biological function – for example, circulating macrophages had a different shape compared with those at different stages of immune attack.
Several colleagues at UTSW are already using the new platform in their research, Drs. Daetwyler and Fiolka said. The control software for the new microscope is open source, allowing researchers elsewhere to customize it for their needs.
“A broader understanding of biological processes across scales will impact our knowledge about many diseases,” Dr. Daetwyler said. “This includes cancer progression, metastasis, developmental disorders, systemic diseases, and cardiovascular diseases, to name just a few.”
Other UTSW researchers who contributed to this study include Gaudenz Danuser, Ph.D., Chair and Professor of the Lyda Hill Department of Bioinformatics; Rolf Brekken, Ph.D., Professor of Surgery, in the Hamon Center for Therapeutic Oncology Research, and of Pharmacology; Felix Zhou, Ph.D., and Dagan Segal, Ph.D., Instructors in the Lyda Hill Department of Bioinformatics; Jill Westcott, Ph.D., research scientist; and Hanieh Mazloom-Farsibaf, Ph.D., and Bingying Chen, Ph.D., postdoctoral researchers.
Dr. Fiolka is a member of the Harold C. Simmons Comprehensive Cancer Center.
This study was funded by grants from the Swiss National Science Foundation (191347), the National Institute of General Medical Sciences (R35 GM133522), and the National Cancer Institute (U54 CA268072 and K99CA270285).
